
SARS-Cov-2 - part 3 - nsp1: A hopefully more detailed analysis of the cellular saboteur
My German cartesian mind (here we go with stereotypes) asks me to start with the first protein expressed by the viral genome of SARS-Cov-2, nsp1. That is, by far, not the low hanging fruit. But it is interesting to learn more about the roles of the different constituents of the virus. Nsp1 actually has also a few very interesting roles to facilitate the viral lifecycle in its host environment.
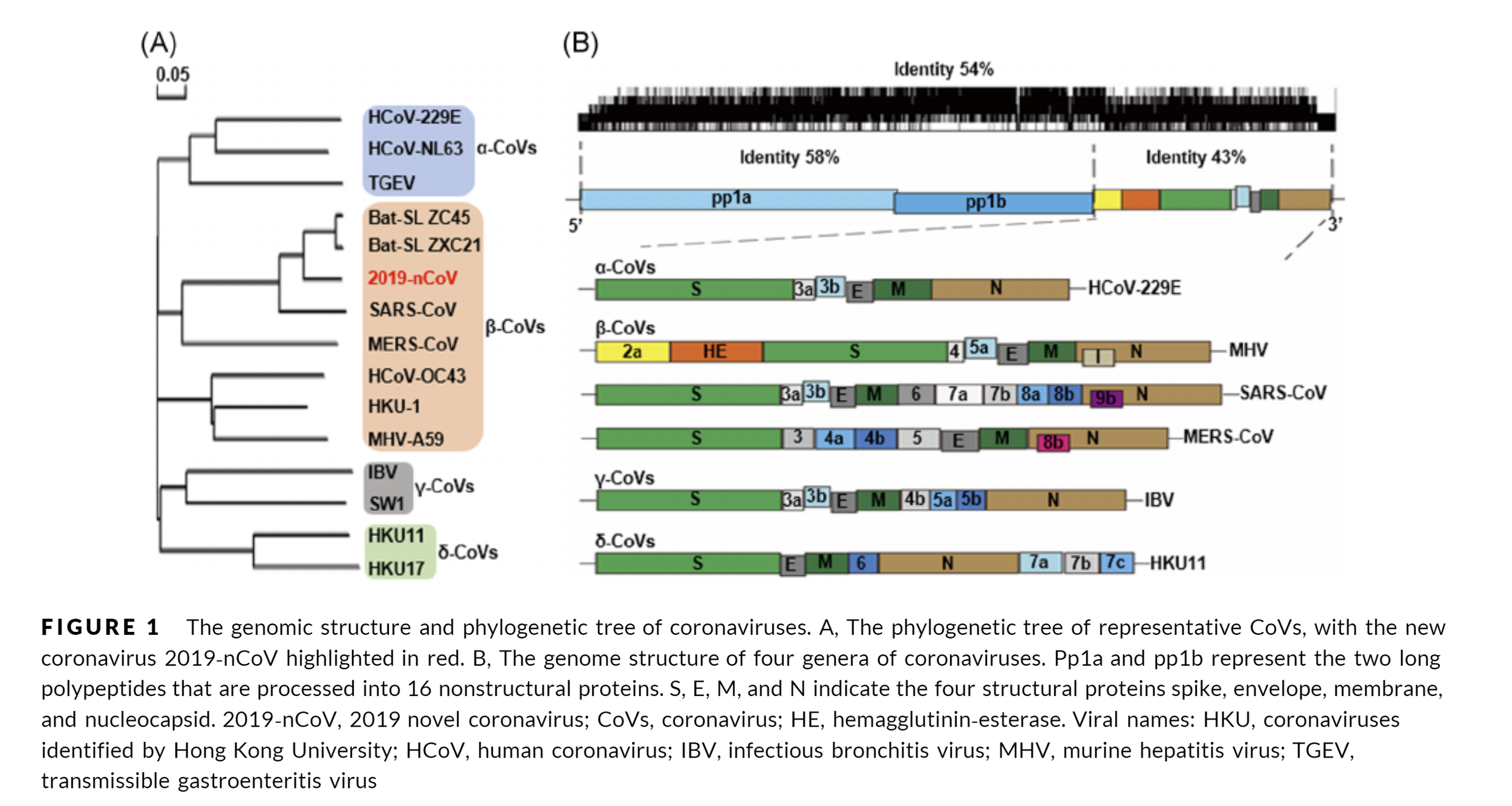
SARS-CoV-2 - part 2 - From the viral genome to protein structures
The bread and butter of every structure based drug design or drug ID effort is the need for 3D structural information of a target of interest related to the phenotypic outcome we want to alter. In the case of SARS-Cov-2 several experimental structures are already available in the RCSB PDB. Other a large scale fragment screens from the Diamond Light Source are slowly becoming available (available in the 3decision protease project for now) on public resources likes the RCSB PDB. Popular homology modelling services also already started to provide several high quality models for some of the yet not resolved structures of the viral proteome.
But let’s first have a look what the viral genome actually contains and what are currently pursued efforts.

SARS-CoV-2 - part 1 - Thriving for a systematic target and hit ID effort
Gabriella outlined in previous posts (part I & part II) a few things about SARS-Cov-2. Back at the time the crisis was still far away from our daily lives but things have changed dramatically during the last weeks. After our initial communication to make 3decision.discngine.cloud freely available for all Covid-19 related collaboration projects we were contacted independently by several volunteers from academic and industrial groups to collaborate on a global effort. At the mean time a lot of novel initiatives have seen the day and everybody is publishing blog posts (like I am right now), linkedIn articles, chemarxiv or bioarxiv preprints etc. It’s a little bit a scientific wild-west happening right now on a global scale.

Discover 3decision® - Part 2: Creating a collaborative platform for Structure-Based Drug Discovery
As we explained in our previous article, the number of biomolecular structures available for computational analysis is increasing daily, which, for SBDD translates to ever-greater opportunities for discovery.
But wait, is that actually true?

Using Adobe XD for Scientific Web app development - do your mockups
This time around you won’t see any source code. Sorry about that, next time maybe.
Here I’ll talk more about sometimes overlooked aspects when developing applications for R&D in life sciences. So I guess this post might be useful to our competitors or people developing applications within large corporations in the life science & pharma industry … anyway ;)
I’ll take the example of our current 3decision developments. You might have heard or read about 3decision already before. If not, check it out - 3decision .. it’s super cool!


Tethered minimization of small molecules with RDKit
I think that’s my first RDKit post! So reason to celebrate!
Here' I’ll focus on a very nice feature available in Open Source Software and very useful in daily structure based but even ligand based drug design tasks.
The problem
How can I dock small molecules into a receptor and avoid as much as possible the “what is the right pose” problem?

Building the VMD molfile plugin
A brief walkthrough on how to compile the molfile plugin for VMD in order to use it for programming and integrating into your own code.

3decision 2018.3
-
Added structure collections: Enabling superposition of unrelated structures
-
New on-boarding tutorials
-
Created structure relation
-
Added GPCR annotations
-
Added visualization of ligand contacts
-
Search for in-house compounds
-
Added detailed crystallographic information
-
Enabled post-filtering of search results
-
New RESTful endpoint for data download and structure registration

