Release Notes
What’s new in 3decision®?
For a comprehensive 3decision® feature portfolio, download our Functional description document.
3decision 2.2 | November 2025
This release introduces our new beta frontend—a significant upgrade designed to deliver faster performance, greater responsiveness, and a refined user experience. Whether you're working with large datasets or customizing complex 3D views, the new interface makes every interaction smoother and more efficient.
While numerous smaller improvements have been made across the platform, here are the most impactful changes. Check them out in the video and release notes below!
Storyboards: Shareable 3D Sessions
They say “a picture is worth a thousand words” — but in structure-based projects, an interactive 3D view can be worth even more.
With Storyboards, you can move beyond static screenshots and create dynamic, shareable narratives that bring your structures to life. Instead of sending isolated PowerPoint slides, PyMOL sessions, or lengthy explanations, Storyboards let you:
Capture multiple 3D scenes with custom views, annotations, and highlights
Add descriptive text to explain context, key interactions, or design rationale
Share a complete visual story with your team—so everyone sees not just the data, but the reasoning behind it
Whether you use Storyboards to save your own work—after spending time perfecting a superposition and customizing views—or as a communication tool with colleagues, or even as support material for a team meeting, this feature makes your insights clear, visual, and interactive. It’s the easiest way to preserve the details that matter and ensure your message gets across.
Click on the GIF to zoom in: 3decision Storyboard feature
New Advanced 3D Viewer
Visualizing structures is now more powerful and flexible than ever. The upgraded 3D viewer allows you to:
Add/hide custom representations (e.g., highlight key residues in green)
Hide individual chains and objects
Add labels and annotations
Generate high-resolution images
Select neighboring atoms or residues within a specified distance
Customizable Search Results Table
The redesigned results page now features a fully customizable structure table (see images below), giving you more control over how you view and manage your data:
Sort structures by any property
Add or remove columns to tailor your view
Filter by author or import date to track in-house data
Add additional search queries for deeper refinement
Click on the image to zoom in: The revamped search result page in the structure view mode (left) and ligand view mode (right)
3decision 2.0 | May, 2025
In this new release, we've achieved two significant milestones: one in Biologics Discovery and another in our collaboration with Chemical Computing Group (CCG). Both milestones add considerable value to the user by enhancing the structure analytics capabilities and the data quality.
Biologics Discovery Milestone
Structure-based biologics projects often require a different set of tools compared to classic small molecule projects, especially for extracting insights on key interactions, binding modes, and structural differences between complexes. To address these needs, we've introduced several new features specifically focused on Biologics Discovery projects, including:
Superposition by chain
Up to now, 3decision has offered the superposition of structures around a binding pocket. With the 3decision 2.0 release, which aims to further support biologics research, we’ve expanded the superposition capability beyond pocket-based alignment to a chain-based one. Biologics researchers often need to align protein chains in order to compare structural arrangements, for example, different antibody binding modes. The new superposition option allows users to manually align specific chains across structures and offers fine control via an optional residue selection.
This new alignment mode is accessible directly in the workspace and when preparing a project. You can thus pre-align your structures by chain and share them within the team, so non-experts can interpret the structural differences more easily and focus directly on relevant regions.
Collaboration around sequence annotations
Scientists working with biologics frequently need to communicate information about specific residues in a protein construct, such as CDR regions, chemical liability motifs, or alternative residue numbering schemes. Whether experimental or predicted, these properties play a crucial role in internal analysis and decision-making.
With the release of 3decision 2.0, users can now register structure residue annotations through the API. These annotations are automatically displayed in the Annotation Browser, where they can be highlighted in 3D at their corresponding residue positions, enhancing visualization and interpretation.
In addition, direct links to the Annotation Browser, highlighting a specific annotation, can now be shared between users.
Discover new features in action in our release video
Click to zoom in: Superposition by chain feature (left) and custom sequence annotations (right) - the new milestone 3decision 2.0 features
CCG Collaboration Milestone
As part of our collaboration with Chemical Computing Group (CCG), we are merging our respective protein structure repositories, 3decision and PSILO, into a unified solution.
With the 3decision 2.0 release, we've achieved a first milestone: PSILO's structure file preprocessing step is now integrated into 3decision's structure import process. This upgrade gives two direct benefits – an enhanced ligand correction and an improved structure file parsing, both resulting in improved data quality for the end user.
3decision 1.10 | August, 2024
PDBx/mmCIF compatibility
In the new release, 3decision enhances its structure repository capabilities by adding support for the PDBx/mmCIF format. As PDBx/mmCIF becomes the new data standard for structural biology, and its ModelCIF extension gains traction within the AI/ML community, this upgrade marks an important milestone.
However, the classical PDB format remains widely used in the industry and is likely to persist for some time, particularly for compatibility with other software. Therefore, 3decision continues to support both and allows users to easily convert structures between the two formats.
In general, structure files (outside of the wwPDB setting) often lack the necessary metadata to fully describe the structure. For example, the description of the ligand’s chemical structure is rarely included in a standardized way, and the same applies to the protein reference for each protein chain. In this new release, 3decision allows users to export enriched PDBx/mmCIF files that contain all metadata provided during the deposit process or after updates. This capability enables 3decision to transform scattered chemistry, biology, and structural data into self-contained files.
Click to zoom in: PDBx/mmCIF compatibility in 3decision
Enhanced Information Browser
3decision’s Information Browser has been restructured to provide users with a more comprehensive and detailed overview of experimental details parsed from the structure header. Depending on the experimental method used, a selection of structure parameters is categorized and displayed in dedicated sections within a new tab called METHODS.
In addition to experimental structures, the headers of ModelCIF structures are also parsed, with modeling parameters and references displayed in a similar manner.
Click to zoom in: Methods tab in 3decision with experimental details
3decision 1.9 | May, 2024
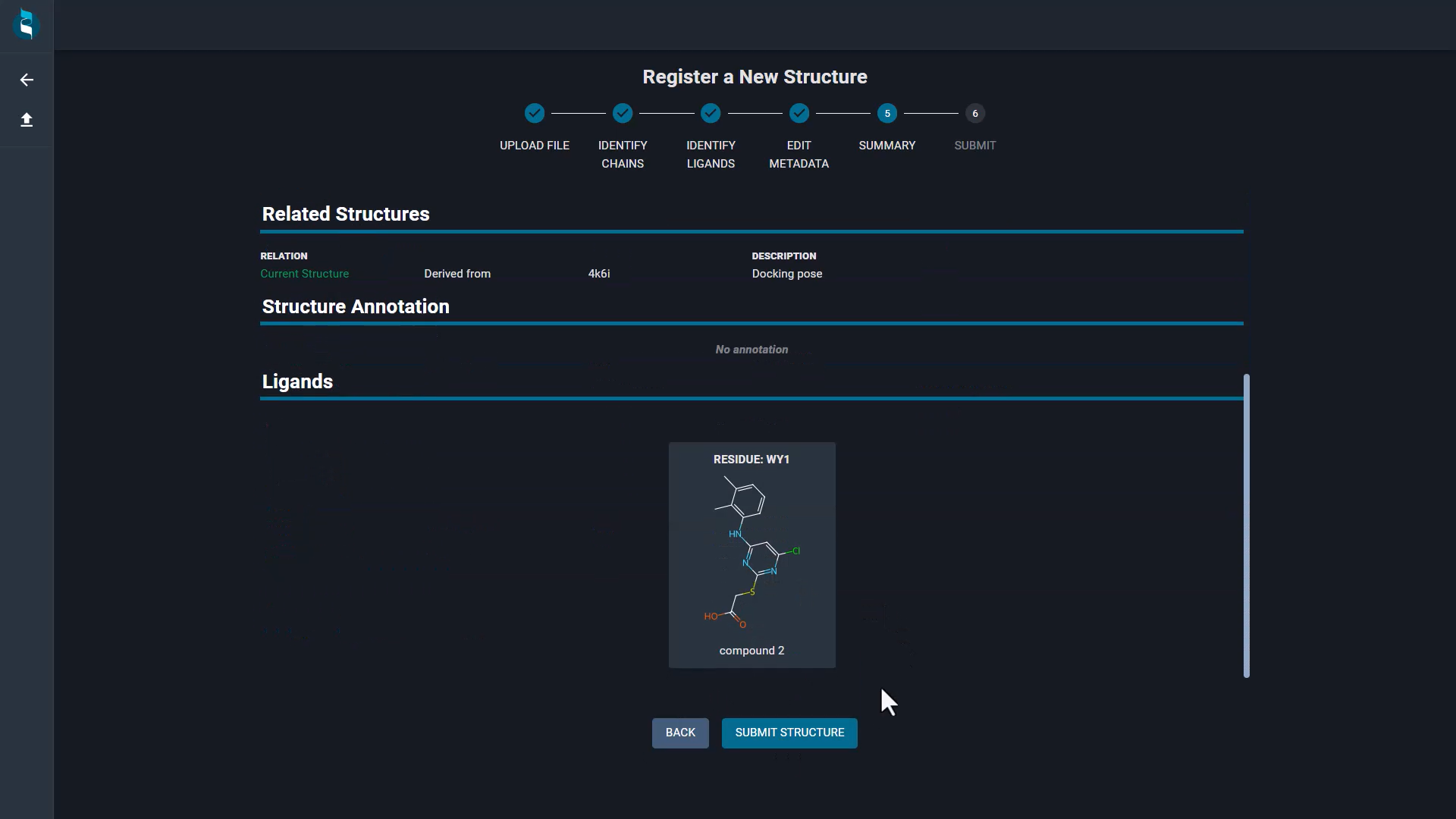
Structure registration from the UI
The latest update to 3decision brings a new feature: Structure Registration from the User Interface (UI). This enhancement is designed to streamline the process of depositing new structures into the 3decision repository, making it more accessible and efficient for all users. Whether they are working with experimental or predicted structures, this new workflow simplifies the registration process, ensuring that users’ data is uploaded efficiently and systematically processed.
The registration process consists of six simple steps, guiding users through each stage of structure checks and association of metadata to the structure with clear instructions. The intuitive design ensures a smooth experience, even for users who are new to the platform. Thanks to this functionality users can enrich the database with experimental and predicted structures that can be shared with the rest of the team in structure-based projects.
3decision 1.8 | January, 2024
AlphaFold Integration in 3decision
3decision incorporates fully annotated AlphaFold (AF) structures of the human proteome into its database, enhancing the depth of available data within the software. This integration of over 23,000 AF structures, complement the pre-existing models in 3decision, sourced from the PDB and the Swiss-Model database, while also accommodating in-house protein structures. The addition of this new dataset can result in a faster and more efficient drug discovery process by providing additional protein structures that can then be used in collaborative projects for analysis and ideation.
Click to Zoom in: AF model in the 3decision 3D Viewer, color-coded by the AlphafoldDB structural confidence score (pLDDT)
On top of the human AF models, users can easily register AF models from other species of interest in just a few clicks. This allows the enrichment of the database with all the relevant information for advancing drug discovery projects. For instance, incorporating structural insights from mouse or rat models may be beneficial in selecting animal models for in vivo drug studies.
Click to Zoom in: Registration page for AF models in 3decision
Upon registration in the 3decision database, AF models undergo the same analysis and processing as any other 3D protein structure deposited in the database. This allows the user to integrate the information provided by these additional structures into their workflows. From basic tasks (like search and visualization) up to advanced analytic functions (for instance, pocket similarity search), AF models are now fully available for supporting 3decision users in their drug discovery efforts.
Click to Zoom in: Comparison between AF model vs experimental
3decision 1.6 | June, 2023
Protein-Ligand Interaction-Based Search
A large amount of structural data on protein-ligand (P-L) interactions provides a knowledge base for scientists working in molecular design. To exploit these data at their best, we developed the interaction search feature that is now available again, after the 3decision back-end revamp.
This systematic structural search allows you to mine and access P-L interaction information which can drive the ideation process of new molecular entities as well as help find putative off-targets through cross-target investigation.
How does the Interaction search feature work?
Interaction search enables you to gather, in a few minutes, systems containing a 3D query pattern of interactions by mining the entire PDB database (and more)
The search is based on a selection of interactions that the user provided as a query system
Interactants and interaction type, interaction length, and orientation tolerance can be refined, offering the user the freedom to define the level of fuzziness of the search.
The search results are ranked based on score and can be automatically superposed in the 3D Viewer.
Click to Zoom in to see interaction search interface and check it out in action in this video
Access the webinar about the drug design idea generation for scaffold hopping with the Interaction Search feature.
Visualization of MTZ Maps
3decision now supports the display of MTZ files. You can upload an mtz. file directly through the structure information widget. The electron density maps are computed on-the-fly, and 2Fo-Fc and Fo-Fc maps are automatically opened in the 3D Viewer.
Click to Zoom in: Visualization of mtz. files in 3decision
3decision 1.5 | April, 2023
Calculation of Protein-Ligand Interactions for Proprietary Structures
When you open a proprietary protein structure in a complex with a ligand in the 3D Viewer, all Protein-Ligand interactions are automatically displayed. Those interactions are calculated and characterized upon registration of the structure in the 3decision database. This feature allows you to easily inspect the ligand binding mode without the necessity of exporting the structure in another program.
Click to Zoom in: Visualization of protein-ligand contacts upon opening a proprietary structure
Bond Order Display
From now on, correct ligand bond orders are displayed in the 3D Viewer, and each ligand gets its representation card for visibility and color management.
Click to Zoom in: Bond order display in the 3D Viewer (right)
Additional Updates:
We have worked on the release of other updates, such as:
Uploading files to a structure or a project is now possible from any file browsing view (Workspace and Project Dashboard)
Project-related files are now accessible directly from the Workspace
Superposition of structures is now enabled from the ligand panel too
3decision 1.4 | February, 2023
Pocket Similarity Search: Global Pocket and Subpocket Search
The search capabilities of 3decision are being extended with the release of the pocket and sub-pocket similarity search, allowing you to mine the structural proteome for global and similar local protein environments.
Usecases:
Identify potential off-targets or search for chemical matter that fits in a similar environment in other structures
Find structures of your target that have a specific conformation of the binding site (e.g. type II binding sites in kinases, closed conformation of HSP90)
How does the subpocket search work?
The user selects a set of amino acids in the binding site then a panel opens allowing to parametrize the query. Once results are back, the user can automatically superpose the hit to the query using the matching residues by simply clicking on a retrieved structure
Click to Zoom in: 3decision sub-pocket search feature. See the video →
Click below to access the Webinar introducing the Subpocket Search feature methodology and use cases
3decision 1.3 | December, 2022
Download and Display of the Public Cryo-EM Maps
In addition to the X-Ray and NMR electron density maps that are automatically retrieved from RCSB PDB, you can now open and visualize the maps of Cryo-EM structures coming from EMDB.
Click to Zoom in: Electron density Cryo-EM map display
Public Access to 3decision User Guide
The user guide of 3decision is publicly available via this link. Moreover, the 3decision users can access the Guide via the application using the “Help” button.
3decision 1.2 | September, 2022
Project Dashboard and Reference Structure Orientation
The Project Dashboard is a centralized view of your project structures, ligands, and files. It summarizes all data associated to your project. It allows you to accomplish following tasks:
Isolate and share stuctures only with a subset of users
Keep an organized list of structures of interest for a project
Define a reference structure and viewpoint to standardize views
In its simplest form, a project is a list of structures of interest a user can share with other users.
Users can set the reference and default viewpoint for a project once and for all. To do so, user can select the pocket of interest, set it as a reference and every structure in the project will be oriented the same way
Click to Zoom in: Reference Structure Setup
Display Protein-Protein Interaction in the 3D Viewer
This 3decision feature allows you to visualize protein-protein interactions (PPIs) and compare them with interactions in other structures.
You are able to display contacts between protein chains in the 3D Viewer. The contacts are generated on the fly using NGL and are not registered in the database.
Click to Zoom in: Protein-protein interactions display
3decision 1.1 | April, 2022
Annotation Browser
The Annotation Browser allows to map structure and sequence annotations onto a 3D structure. This feature also allows you to add and visualize your own annotations.
This layout combines, on the left side, sequence annotations which have interactive selection with the 3D Viewer, on the right side. Each protein chain in the structure is displayed on the left under its Uniprot identifier. The full sequence and a list of annotations, coming from various sources are displayed.
Usecases :
Highlight a CDR motif on an antibody structure
Examine which part of the canonical sequence is covered by the resolved structure
Localize a residue on a 3D structure using a protein-specific numbering scheme (e.g. Ballesteros–Weinstein for GPCRs)
Add a new sequence annotation (e.g. result of a point-mutation) 3decision 1.1
Click on the video to explore the Annotation Search Feature
3decision 1.0 | January, 2022
Highlight Mode
To ease the multi-structural analysis, this new feature enables users to spot the differences and/or common parts of the superposed binding sites in the blink of an eye — one more tool to further support easy pocket comparison.
Click to Zoom in: 3decision highlight mode 💡
Pocket Browser
The Pocket Browser allows the user to explore all pockets and ligands of a given protein structure. Both ligand-binding sites and ligand-agnostic pockets are identified and characterized by fpocket when a structure is registered in 3decision. Each pocket is characterized based on the properties such as druggability, polarity, volume, hydrophobicity, and charge.
There are two types of pockets:
explicit pockets, when the pocket is defined by the presence of a ligand (represented by squares in the legacy version)
implicit pockets are pockets detected by fpocket (represented by circles in the legacy version)
The Pocket Browser table is interactive with the 3D structural information. Click on the pockets to navigate in the structure through the different pockets.
Easier Comparison of Grids & Electron Density Maps on Superposed Structures
When superposing two structures and fetching (or importing) electron density maps, they are automatically re-oriented along with the superposed structures.
User Groups and Privileges
It is now possible to organize team members into User Groups to control data access and actions and to easily share content. Each of these actions is well defined through the completely new set of features, covered by the term “Privilege Management.” Privileges are introduced to allow users to do particular actions, such as to view/create/update/remove specific structures or projects.
Search / Advanced Search and Speed Improvement
3decision®'s knowledge-base can be explored through various search queries:
Text search (Free text, UniProt code, Gene name, Protein family, PDB code, Ligand ID, etc.)
Chemistry search
Exact match
Similarity search
Substructure
User can combine text and chemistry searches by adding each of the two partial search in the right panel.
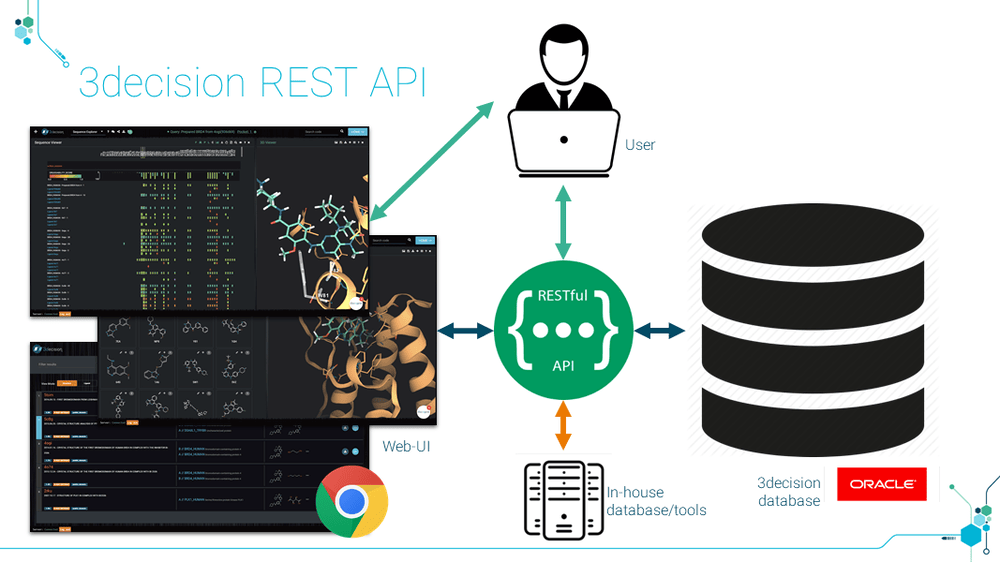
3decision public REST API release allows management of
Structures: registration (bulk)/ Removal, retrieval of files and associated ligands.
Metadata: projects, annotations, crystallographic parameters
Associated files: .ccp4, .mtz, .pdf, .sdf
Project management
The complete list of REST endpoints can be shared on-demand.
3decision has a unique structure registration and analysis process through API endpoints. It covers:
Structure-to-sequence mapping
Ligand mapping
Pocket analysis
Pocket feature analysis
Ligand to pocket mapping
Protein-ligand contact analysis
Click to Zoom in: 3decision Rest API
The first version of 3decision has been released in 2018 and gradually upgraded with various features. In 2022, the backend of the 3decision solution has been revamped to improve customer experience and enhance the performance. The release notes are based on the new version.