Starting with 3decision®: Allosteric pocket detection in BCR-ABL1 kinase
I recently joined Discngine as a member of the 3decision® team. After my onboarding period and training on the platform, I worked on small projects to put my knowledge into practice. In this article, you will join me on my journey, and I will share what I learned about some of the 3decision® features. In my first project, my goal was to investigate the ability of 3decision® to identify druggable binding sites on a protein surface. To do so, I performed a retrospective analysis of the detection of the allosteric pockets in the ABL1 kinase.
Introduction
Let’s start with a few words about the selected target.
The ABL1 non-receptor tyrosine kinase is an essential enzyme for the regulation of various cellular processes (cell survival, proliferation, oxidative stress, and much more). The chromosomal translocation fuses the part of the breakpoint cluster region (bcr) gene and c-abl gene, producing the oncoprotein BCR-ABL1. It acts as a tyrosine kinase and has been implicated in the pathogenesis of chronic myeloid leukemia (CML).
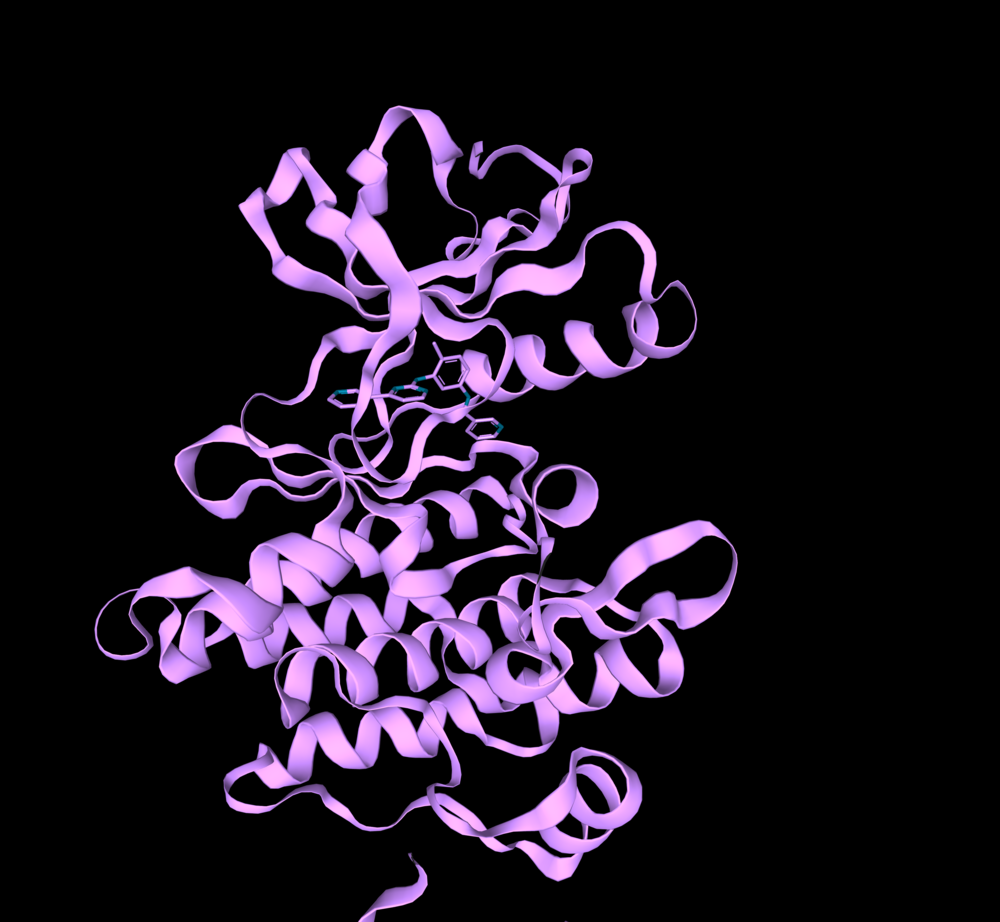
BCR-ABL1 kinase (PDB:1IEP).
Currently, more than 5 US Food and Drug Administration (FDA) drugs targeting the kinase ATP-binding site have been approved against this malignancy. The complication associated with this therapy is the very frequent, clinically acquired resistance that is induced by point mutations at the orthosteric binding site. Such a type of resistance occurs often after just 1-2 years of clinical use. Moreover, the fact that the ATP binding site is highly conserved among the kinome, brings many unwanted reactions to the treated patients due to the insufficient selectivity of the drugs used in the therapy.
To address the lack of selectivity, one of the alternative strategies is to develop novel kinase inhibitors that are targeting allosteric binding sites. The advantages include the achievement of greater selectivity with less off-target effects and the minimization of point mutation-associated drug resistance (although this approach is still under clinical investigation today). The Bcr-Abl is one of the first kinases whose allosteric pocket has been identified and for which the allosteric inhibitor Asciminib (ABL001) showed a biological activity (all of this happened years after the crystal structures I have used in this case study were released).
Recently, pharma company Novartis has been granted the Breakthrough Therapy designation (BTD) by the FDA for the development of Asciminib, which is currently undergoing phase 3 clinical trials for the treatment of CML.
UPDATE: At the end of October 2021, FDA approved asciminib (Scemblix®) for the treatment of chronic myeloid leukemia (CML) in two indications, one of them being patients with T315I mutations.
Here is a short visual summary of kinase inhibitor discovery
Pocket identification
The entire PDB database is registered in 3decision® and updated regularly. Using the text search “abl1” and filtering the date of structure release (2000-2001), I found two crystal structures of BCR-ABL1 in mice (PDB IDs: 1IEP, 1FPU) that had a co-crystalized ligand in the orthosteric binding site. These structures are from the period of the discovery of imatinib – the first-ever FDA approved drug against BCR-ABL1 kinase (the year 2001). I chose to work with them because at that time the allosteric pocket has not yet been determined.
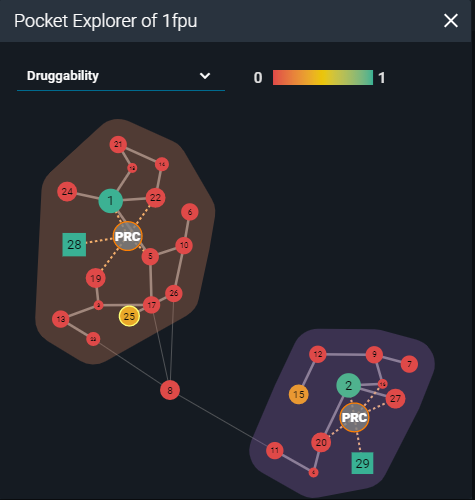
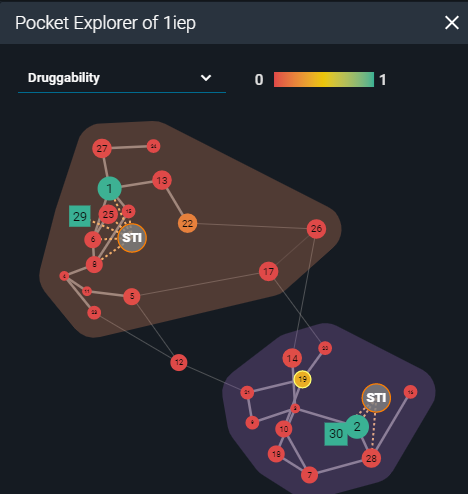
Then I started an analysis of the structural surface. This is possible thanks to the calculations performed by the fpocket cavity detection algorithm, presented in the 3decision Pocket Explorer. Fpocket scans the molecular surface of the protein for pockets across all the structures registered in 3decision. Each pocket is characterized using properties such as druggability, polarity, volume, hydrophobicity, and charge. The Pocket Explorer map visually represents each pocket as a round, or a square shape (depending on the identification method) and each calculated property is color-coded for quicker interpretation. The identified druggable sites can be considered as a starting point for in-silico structure-based ligand design.
Representation of pocket explorer map with scored cavities (PDB:1FPU).
Representation of pocket explorer map with scored cavities (PDB:1IEP).
On the Pocket map of the 2 analyzed structures I worked on, there was one additional cavity with a druggable score above 0.3, besides the one already occupied by the ligand: pocket number 25 in 1FPU and pocket number 19 in 1IEP (druggability scores: 0.36; score range 0-1). I superposed them to compare the pocket locations on their surfaces. The druggable pockets in both structures occupied the same space on the 3D structures. They were located on the C-terminal lobe of the kinase domain, far from the active site. The residue-based calculated properties implied that this pocket has a high hydrophobic character and a low score of polarity and charges. By visual inspection of the pocket shape, one may conclude that despite the low volume, it results to be deep enough to potentially accommodate a ligand.
Superposition of “druggable” pockets in the two analyzed structures.
I then compared my findings with the literature. What 3decision® suggested as a druggable site in the structures of ABL1 from the year 2001 turned out to be the “myristoyl-binding pocket”, discovered a couple of years later (in 2003). The complex of an ABL1 kinase and an endogenous myristoyl group was determined to contribute to the auto-inhibition of the ABL1 kinase (the regulatory mechanism lost in the BCR-ABL1), and it is the location where the novel inhibitor Asciminib binds. The recently resolved crystal structure of ABL1 with Asciminib (PDB ID: 5MO4) shows how is this binding site accommodates the ligand, which is proven to be biologically active.
The allosteric binding site of BCR-ABL1 with novel inhibitor Asciminib (PDB: 5MO4).
Summary
The detection of ABL1 kinase allosteric pocket in a retrospective way was great practice for me to learn some of the features of 3decision®. The Pocket Explorer enables 3decision to identify and quickly characterize all cavities on the protein surface. Moreover, each pocket is assigned a druggability score, which helps the user estimate the druggability of a target. This method can be very useful for structural analysis, especially in the very first steps of structure-based drug discovery.
The next steps in my journey would be to continue to explore more features in 3decision, and I am very excited to share my new findings here!
In the meantime, feel free to check out 3decision® directly on the cloud!
References: